Body fat composition and miR-378 expression profiling in patients with type 1 diabetes
Article information
Abstract
Purpose
Type 1 diabetes (T1D) is an autoimmune disease that involves genetic, epigenetic, and environmental factors. Change in body composition is a potential mechanism for explaining the increased incidence of T1D. Micro RNA-378 (miRNA-378) is a positive regulator of adipogenesis that has yet to be studied in such patients. This study aims to evaluate the miRNA-378 expression profile in peripheral mononuclear cells of T1D patients and controls and to determine its possible association with levels of body fat, interleukin-6 (IL-6), and tumor necrosis factor alpha (TNF-α).
Methods
Twenty-four T1D subjects and 20 controls under 18 years of age without autoimmune diseases were studied. miRNA-378 expression profile was determined by TaqMan probes. Body composition was determined by multifrequency bioimpedance. IL-6 and TNF-α serum levels were determined by LUMINEX. AntiGAD65, anti-IA2, and anti-ZnT8 antibodies were quantified in serum by enzyme immunoassays. Statistical significance was considered P<0.05.
Results
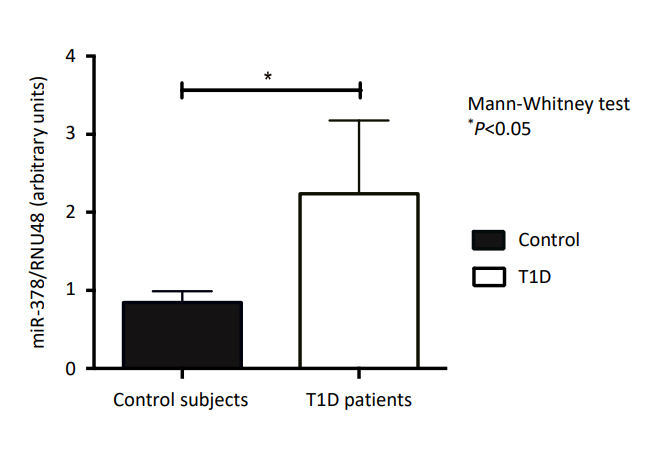
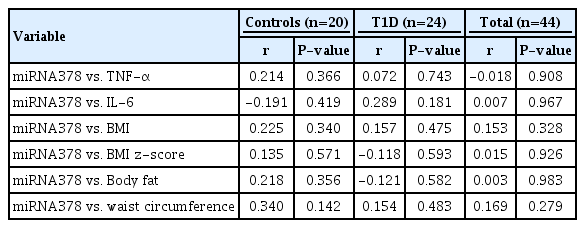
Similar body mass index and body fat (kg) were observed between the T1D and control subjects (P=0.55 and P=0.69, respectively). The miRNA-378 expression profile was significantly higher in T1D patients compared with the controls (P<0.05). Lower miRNA-378 expression in prepubertal controls was observed compared to pubertal controls, prepubertal T1D, and pubertal T1D (P<0.05). AntiGAD65, AntilA2, and AntiZnT8 were positively correlated with miRNA-378 (P=0.002, P=0.053, and P=0.007). No statistically significant correlation was observed between miRNA-378 expression and IL-6, TNF-α, or body fat.
Conclusions
Elevated miRNA-378 expression in T1D patients compared with controls is linked to pubertal stage but is not associated with proinflammatory status or body composition.
Introduction
Type 1 diabetes (T1D) is a chronic autoimmune disease of multifactorial etiology that results in destruction of pancreatic β cells [1]. Since the 1980s, the incidence of T1D has increased globally [2]. Through the DiaMond Study (Diabetes Mondiale Project Group) published in 2000, Chile was classified as a country with a low incidence of T1D with 2.36 new cases per year per 100,000 inhabitants of Santiago, Chile's capital city, under 15 years of age (data obtained between 1990 and 1992) [3]. Between 1994 and 2000, however, the incidence of T1D in Chile increased to 4.5 per 100,000 inhabitants (<15 years) of Santiago [4]. Together, the increase in worldwide T1D incidence and the reduced number of genotypes that confer an increased T1D risk suggest that the environment is a predisposing factor to T1D. Potential mechanisms may include modifications in gene expression due to epigenetic changes [5].
Over the last 3 decades, there has been an increasing percentage of obesity in both developed and developing countries [6]. In Chile, it has been reported that 50% of the child population is overweight or obese [7]. The global increase in prevalence of overweight and obesity may result in epigenetic changes due to excessive accumulation of adipose tissue and subsequent infiltration of macrophages in production of proinflammatory cytokines [8]. Several cytokines that are released by adipose tissue, such as tumor necrosis factor alpha (TNF-α) and interleukin-6 (IL-6), have been proposed along with other mediators of inflammation to being related to generation of both T1D and type 2 diabetes [9].
There is increasing evidence linking miRNAs to autoimmune pathologies [10]. Other studies have shown that certain miRNA activity is related to adipose tissue development and function [11]. In this context, miR-378 has been reported to be upregulated in human adipocytes treated with IL-6, TNF-α, and leptin [12]. This miRNA is coded by an intron of the peroxisome proliferatoractivated receptor gamma coactivator 1β gene (PPARGC1β), counterbalancing the metabolic actions of peroxisome proliferator-activated receptor gamma coactivator 1β (PGC1β). Micro RNA-378 (miRNA-378) has also been identified as integral to the human regulatory system that controls systemic energy homeostasis and total oxidative capacity of insulin target tissues [13]. Furthermore, it is now apparent that miR-378 plays a role in several biological processes, such as cancer metastasis, osteoblast differentiation, and adipogenesis [14,15].
Given that Chile has been facing concurrent increases in both T1D and childhood obesity, Chile provides a suitable environment in which to further study the mechanisms related to the association between excess body fat and pathogenesis of T1D. Therefore, we sought to evaluate the expression profile of miRNA-378, levels of IL-6 and TNF-α, and their possible association with body fat in T1D Chilean patients.
Materials and methods
1. Study participants
In this study, 24 children with T1D from Santiago, Chile (from the Maternal and Child Research Institute [IDIMI], University of Chile) and 20 healthy controls without T1D under 18 years of age were enrolled. Study participants were recruited from the Maternal and Child Research Institute (IDIMI, for its name in Spanish) at the University of Chile. Since sex-specific differences in microRNAs have not been described, the study sample was designed independent of sex. Study participants were also recruited independent of nutritional status and pubertal stage. The primary caregiver or parent of each study subject provided written informed consent, and assent was obtained from all study participants. A survey was conducted that compiled the patient's medical, family, and clinical histories. This study was approved by the Ethics Committees of the Faculty of Medicine and IDIMI of the University of Chile.
2. Anthropometry and body composition analysis
Height was measured to the nearest centimeter using a rigid stadiometer (SECA, Hamburg, Germany). Weight was estimated (without clothing) using a balance (SECA) calibrated at 0.1 kg. Body mass index (BMI) was calculated as weight (kg)/height (m2). Pubertal stage was classified using Tanner staging. Waist circumference was measured to the nearest centimeter using an inelastic tape measure (SECA), considering the smallest circumference between the costal border and the iliac crest. Body composition analysis was performed using multifrequency bioimpedance with InBody S10 equipment (Seoul, Korea). Nutritional status was classified according to the 2006 World Health Organization (WHO) growth charts for children 0 to 5 years of age considering weight-for-length/height, weight-for-age, and length/height-for-age indicators and 2007 WHO growth charts for children less than 5 years of age using height-for-age and BMI-for-age indicators [16].
3. Sample collection blood draws
Blood samples were collected from all study participants in the San Borja Arriarán Hospital. In all cases, 12 mL of blood were obtained through venous puncture.
4. Extraction of mononuclear cells
Each of the 12-mL blood samples was diluted with a saline phosphate buffer (phosphate-buffered saline, PBS) in a 1:1 ratio to facilitate the handling of the sample. Diluted samples were slowly disolved in Ficoll-Hypaque solution (lymphocyte separation medium, 1,077-g/mL density; Cellgro, Manassas, VA, USA) and centrifuged for 30 minutes at 1,840 rpm (410 g) and 4℃. After centrifugation, the peripheral blood mononuclear cells were transferred to a new tube and washed twice with PBS 1× through 10 minutes of centrifugation at 2,400 rpm (730 g). Finally, the obtained cells were resuspended in RPMI-1640 (medium without glucose) (Gibco, Invitrogen, Waltham, MA, USA) and then counted using trypan blue stain (Trypan Blue Stain 0,4%; Gibco, Invitrogen, USA) under Neubauer cameras.
5. Total RNA extraction
Total RNA extraction was performed with the TRIzol method (Invitrogen). Purity by lack of phenol or chloroform contamination was determined by the absorbance ratio A260/280 and A260/230. RNA integrity was evaluated through the relationship between the 28S and 18S ribosomal bands and subsequently quantified by absorbance spectrometry (Infinite NanoQuant M200 Absorbance Microplate Reader, TECAN, Männedorf, Switzerland). Finally, cDNA was synthesized from 10 ng of total RNA using the TaqMan small RNA assay (Applied Biosystems by Life Technology, Foster City, CA, USA).
6. MicroRNA expression
RT-stem loop (Applied Biosystem) specific for miR-378 was used. Expression levels were determined with TaqMan MGB probes and TaqMan Universal PCR Master Mix II (2x) not UNG, in the Agilent Mx3005P (Agilent Technologies). Expression levels of miR-378 were normalized to RNU48.
7. Serological analysis
Anti-GAD65, anti-IA2, and anti-ZnT8 antibodies were quantified in serum by enzyme immunoassay (ELISA) using commercially available kits Medizym anti-IA2, Medizym antiGAD, and Medizym anti-ZnT8 (Medipan GmbH, Berlin, Germany) according to the manufacturer's instructions for both T1D and the controls. IL-6 and TNF-α levels were determined by LUMINEX assay.
8. Statistical analysis
We used the program REST (Relative Expression Software Tool) to analyze Quantitative PCR values. All subsequent calculations were performed using the software packages SPSS ver. 15.0.1 (SPSS Inc., Chicago, IL, USA) and GraphPad Prism version 5 (GraphPad Software, Inc., San Diego CA, USA). The Shapiro-Wilk normality test was performed to determine the distribution of the variables. To determine if there were differences between control subjects and T1D patients regarding age, BMI, body fat level, waist circumference, and inflammation profile, the Mann-Whitney test was used. To determine if there was an association between miRNA expression, body fat level, inflammation profile, and autoimmunity profile, the KruskalWallis test followed by post hoc Dunns was used. To determine if there was a correlation between miRNA expression, body fat level, inflammation profile, and autoimmunity profile, the Spearman correlation was used. Statistical significance was considered P<0.05.
Results
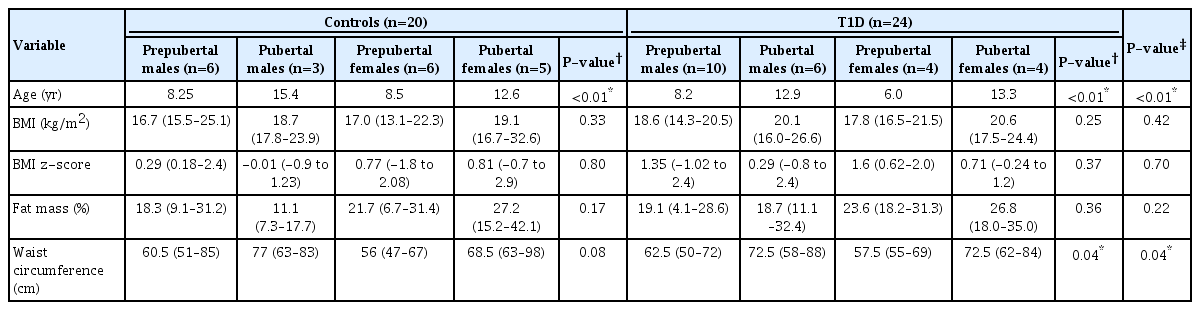
Table 1 shows the clinical, immunological, and inflammatory characteristics of both the T1D and control groups. Nutritional status, body fat (%), IL-6 (pg/mL), and TNF-α (pg/mL) was similar between the groups (Table 1). Categorizing subjects by sex and pubertal stage resulted in a higher body fat percentage in pubertal girls even though there was no significant difference (Table 2). No significant differences were observed in BMI and z-score BMI between the groups. Waist circumference was statistically significantly higher in the pubertal groups compared to the prepubertal groups (Table 2).
The correlation analysis between inflammatory cytokines and anthropometric parameters is shown in Table 3. Significant correlations were observed for TNF-α and IL-6 with BMI and body fat in the total study sample but not in the stratified analysis by group (T1D vs. controls).

Correlation analysis between inflammatory cytokines level vs. BMI, body fat %, and waist circumference of controls and T1D patients
Fig. 1 shows the relative expression of miR-378 in T1D patients and controls. A higher expression of miR-378 was observed in those with T1D compared to the controls (P<0.05).
Fig. 2 shows the relative expression of miR-378 in those with T1D and the controls by pubertal stage. A lower expression of miR-378 was observed in prepubertal controls in comparison to pubertal controls, prepubertal T1D, and pubertal T1D (P<0.05).
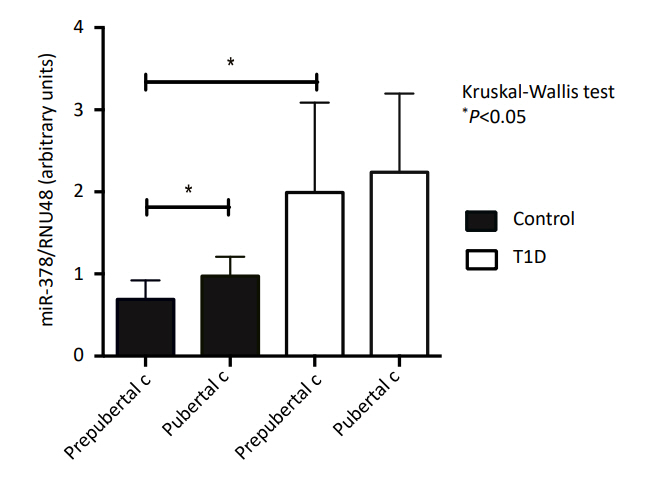
Expression of micro RNA-378 (miRNA-378) in controls and type 1 diabetes (T1D) patients grouped according to pubertal stage.
Fig. 3 shows the correlations between miRNA-378 expression level and autoantibody concentration in those with T1D. Both AntiGAD65 and AntiZnT8 antibodies were statistically significantly positively correlated with miRNA-378. No statistically significant correlation was observed between the AntiIA2 antibody and miRNA-378.
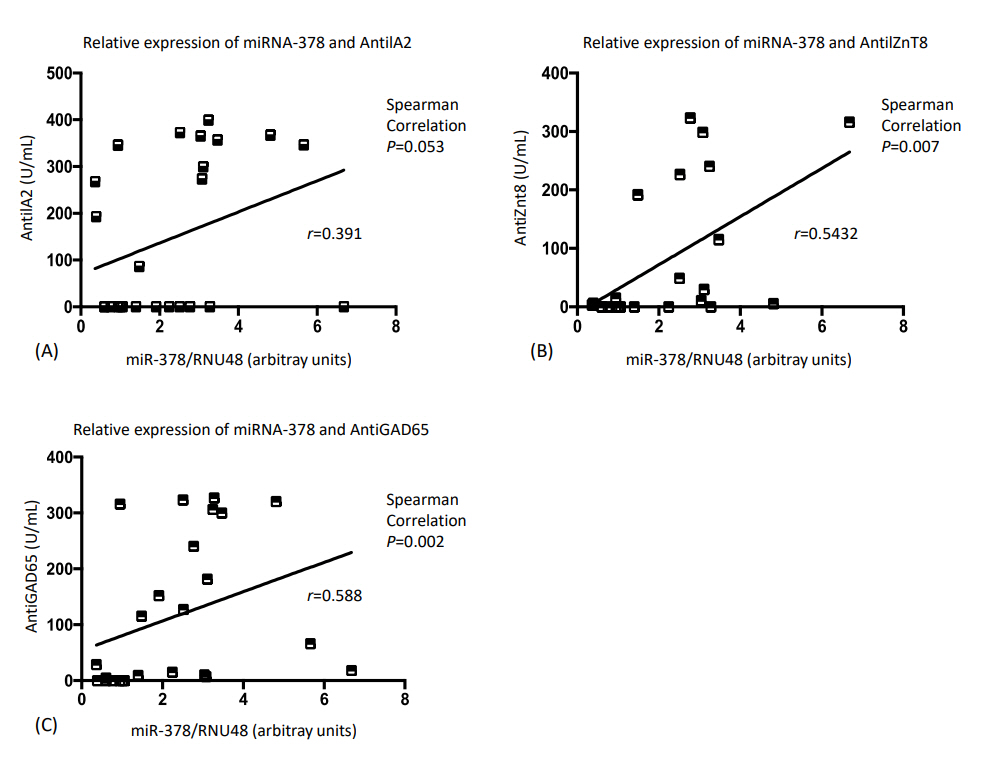
Correlation analysis between micro RNA-378 (miRNA-378) relative expression and autoantibodies level of type 1 diabetes patients.
Finally, Table 4 summarizes the correlations between cytokines and body composition related to miR-378 expression. However, no statistically significant correlations were observed among these parameters.
Discussion
To the best of our knowledge, this is the first study of miR378 expression in T1D juvenile patients. Despite the previously described association between miR-378 and adipose tissue, we did not find associations between miRNA-378 with proinflammatory states or body composition even though we found that miRNA-378 was greater in those with T1D compared to the controls as well as among all subjects undergoing puberty. Furthermore, while the overall study sample had a high prevalence of overweight, this study also found no statistically significant differences in nutritional status (BMI z-score and percent body fat) of children with T1D and the controls.
In recent years, an increase in overweight T1D patients has been described. One study performed by Flechtner-Mors et al. [17] with 18,382 T1D children and teenagers from Germany and Austria showed that, between 2000 and 2006, there was an increase in obesity and overweight prevalence in both girls and boys with T1D. In addition, Dubose et al. [18] developed an investigation including 32,936 children from Germany, Austria, and the United States showing that, according to WHO classification criteria, 24% of patients are overweight and 12% are obese. In contrast, a study by Mosso et al. [19] of 30 T1D Chilean juvenile patients found that only 17% were overweight. The study of Gregory et al. [20] showed a higher body fat percentage in T1D pubertal girls compared to T1D prepubertal subjects and T1D pubertal boys.
Several publications have shown a progressive increase of body fat during puberty compared to the prepuberty stage [20,21]. Comparing those with T1D to those without T1D, weight gain during puberty tends to be higher in those with T1D in both sexes [22,23] and only in girls without T1D [17,19]. The greater weight gain in those with T1D may be explained as undesirable consequence of subcutaneous insulin treatment [23,24] that is known to have a double inhibitor effect of lipolysis and as a lipogenesis stimulator, promoting fat accumulation. This situation is increased during puberty due to higher insulin requirements caused by both higher anabolism and lower insulin sensibility [25].
In our study, no significantly higher BMI or body fat % was observed in T1D patients compared to controls when the analysis was stratified by age and pubertal stage. While we observed a higher waist circumference in pubertal T1D and controls, this was expected considering that weight, and possibly height, are likely to increase during puberty.
A limitation of our study is that we measured total body fat and not the distribution of body fat. Research conducted in El Cairo [26] also found that those with T1D had higher total body and abdominal fat compared to the controls. Taken together, our results are consistent with those by Paulino et al. [27] who also found no significant differences in BMI z-score or body fat % by group (with or without T1D) even though they found a higher waist circumference in T1D patients compared to controls. This fat distribution has been related to glucose control in teenagers, and the excess of adipose tissue has been linked to cardiovascular risk factors in T1D [28].
Our study showed similar inflammatory cytokine levels in diabetic patients compared to controls, which is in agreement with the literature since most T1D patients considered in this study had adequate metabolic control (glycosylated hemoglobin<7.5%) and were diagnosed less than 1 year prior. Recently, Alnek et al. [29], in concordance with our study, found elevated level of IL-1β but not IL-6 or TNF-α in T1D children.
In our study, miR-378 overexpression was observed in T1D patients compared to control patients. However, mi-378 expression was not statistically significantly correlated with percent body fat or inflammatory cytokine level. This may be explained by the similar inflammatory cytokines levels, nutritional status, and body fat of the T1D patients and the controls recruited for this study.
Carrer et al. [13] demonstrated that mice genetically lacking miR-378 are resistant to obesity induced by a high fat diet. Therefore, a reduction in fat mass deposits was observed as well as a diminution of adipocyte size, raising the possibility that this miRNA is necessary for efficient hypertrophy and lipid absorption in adipocytes. miR-378 counteracts the metabolic actions of PGC-1β [13]. PGC-1β acts in conjunction with transcription factors such as estrogen-related receptors (ERRα, ERRβy, and ERRγ), activating mitochondrial biogenesis, fatty acids oxidation, glucose uptake, and gluconeogenesis in several tissues [30].
Studies have shown that diabetic subjects with insulin resistance or obesity have decreased mitochondrial capacity in skeletal muscle that is linked to low PGC-α and PGC-1β expression and, therefore, to deregulation of both mitochondrial genes and their subsequent enzymatic activity [31,32]. miRNA-378 is known to be highly induced during adipogenesis. A study performed by Xu et al. [12], where human mature adipocytes were treated with TNF-α, IL-6, leptin, and resistin, showed miR-378 overexpression in adipocytes treated with TNF-α, IL-6, and leptin but not with resistin. Ishida et al. [33] found that miRNA-378 represses the adiponectin gene in human adipocytes that in turn likely reduces adiponectin expression. In addition, Gerin et al. [34] demonstrated that miRNA378/378* overexpression during adipogenesis increases triacylglycerol accumulation in adipocytes due to the increase of de novo lipogenesis. Therefore, we hypothesize that T1D, as described for other miRNAs [35], may contribute to miRNA-378 overexpression with consequent negative regulation of PGC-1β/ERRs and its target genes. Recent studies have reported that epigenetic factors such as miRNAs play a key role in pathogenesis of T1D, primarily affecting functioning of the immune system and apoptosis of pancreatic β cells [35].
When T1D patients and controls were categorized by pubertal stage, we observed lower miRNA-378 expression in all prepubertal subjects compared to pubertal subjects, suggesting a puberty effect on miRNA-378 expression. Given that miRNA-378 is necessary for adipose tissue hypertrophy [36], it can be explained by the lower insulin sensitivity and increase in fat mass that occurs during puberty [20,21,23]. The higher miR378 expression we found in T1D patients may be related to the greater gain in fat mass during puberty that has been previously observed in diabetic patients [17,19,22,23]. It can also be linked to Wilkin’s “Accelerator Hypothesis,” which indicates that adiposity responsible for insulin resistance could increase betacell demand and induce apoptosis with autoantigen release, accelerating autoimmune damage in genetically predisposed subjects [37].
miR-378 is related to PGC-1β, a coactivator of ERRs. ERR expression and its role in pubertal development have yet to be fully defined. A recent study [38] demonstrated that ERRγ is expressed in mouse Leydig cells, increases synthesis of testicular testosterone, and regulates steroidogenic genes expression, which could link it to pubertal development.
Our study showed positive correlations between autoantibodies and miR-378 relative expression that was only statistically significant for AntiGAD65 and AntiZnT8. Several studies have compared the expression profiles of miRNAs with those of autoantibodies in human subjects with T1D. Salas-Pérez et al. [39] observed decreases in miR-21a and miR-93 expression in T1D children compared to healthy controls but found no correlation between expression of these miRNA and autoantibody profiles. In contrast, the study by Sebastiani et al. [40] is consistent with our findings in which greater expression of miR-326 in T1D subjects was positively correlated with AntiGAD65 and AntiIA2 autoantibodies.
Taken together, the finding indicate that microRNA may have a modulating role in the autoimmune response of T1D patients. Further studies are necessary to determine if the differential miR-378 expression in T1D patients compared to that of healthy controls is due to an event after or before T1D onset–the latter of which could position miR-378 as a potential biomarker for T1D.
Notes
Ethical statement
All procedures followed were in accordance with the ethical standards of the responsible committee on human experimentation (institutional and national) and with the Helsinki Declaration of 1975, as revised in 2008. This study was approved by the Ethics Committees of the Faculty of Medicine and IDIMI of the University of Chile (IRB Nº 053/2014). Informed consent was obtained from all patients included in the study.
Conflicts of interest
No potential conflict of interest relevant to this article was reported.
Acknowledgements
We thank all the participants for their cooperation. This project was supported by FONDECYT Grant 1130240.