Genetic aspects of type 1 diabetes
Article information
Abstract
Type 1 diabetes mellitus (T1DM) is characterized by autoimmune destruction of pancreatic beta-cells in genetically predisposed individuals, eventually resulting in severe insulin deficiency. It is the most common form of diabetes in children and adolescents. Genetic susceptibility plays a crucial role in development of T1DM. The human leukocyte antigen complex plays a key role in the pathogenesis of T1DM. Furthermore, genome-wide association studies and linkage analysis have recently made a significant contribution to current knowledge relative to the impact of genetics on T1DM development and progression. This review focuses on current knowledge of genetics as a pathogenesis for T1DM. It also discusses mechanisms by which genes influence the risk of developing T1DM as well as the clinical and research applications of genetic risk scores in T1DM.
Introduction
Type 1 diabetes mellitus (T1DM) is an autoimmune disease that usually occurs in children and adolescents. T1DM occurs because of insulin deficiency that results from T-cell mediated destruction of insulin producing pancreatic beta-cells [1]. The incidence of T1DM is extremely variable among ethnic groups [2]. Although its incidence in children younger than age 15 has increased over the past 2 decades in Korea, the annual incidence in Asian countries (<5 in every 100,000 individuals), is much lower compared to Europe and other Western countries [3]. The ethnic differences in T1DM incidence in children can be attributed to environmental and genetic risk factors.
The current consensus for the development of T1DM is based on the Eisenbarth T1DM etiopathogenesis model [4]. Activation of T- and B-cell autoimmune responses against beta cells is triggered by environmental factors, such as infection and toxins in genetically predisposed individuals, resulting in beta cell destruction of the pancreas [5]. Approximately 90% of patients with T1DM, have circulating auto-antibodies against islet cell auto-antigens (type 1A) of the pancreas. In addition, approximately 10% of newly diagnosed T1DM patients are auto-antibody negative (type 1B) [6].
In the past 10 years, nationwide and international research consortiums and networks have conducted genome-wide association studies (GWAS) and linkage analysis of T1DM. These include the Type 1 Diabetes Genetic Consortium [7], Diabetes Autoimmunity Study in the Young (DAISY) [8], TrialNet Pathway to Prevention study [9], BABYDIAB [10], Environmental Determinants of Diabetes in the Young [11], and the Wellcome Trust Case Control Consortium (WTCCC) [12]. With the advent of genomic screening, more than 50 non-human leukocyte antigen (HLA) regions as well as HLA genes have been identified in T1DM [13]. This review focuses on current knowledge of genetics as a basis for T1DM and the mechanisms by which genes influence the risk of developing T1DM.
Genetic susceptibility in type 1 diabetes
Genetic susceptibility is one of the major causes of T1DM. Many studies have reported that up to 50% of risk factors for T1DM are hereditary [13]. In the general population, the lifetime risk for developing T1DM is approximately 0.4%; however, relatives of T1DM patients have a higher risk of developing the condition. The identical twin concordance rate for T1DM is 25%–50%, while it is only 6% for dizygotic twins, approximately 6%–7% for siblings, 1.3%–4% for children of female patients, and 6%–9% for children of male patients [14-18]. These concordance rates emphasize the significance of genetics in T1DM.
Human leukocyte antigen
HLA complex plays a critical role in the pathogenesis of T1DM. The association between T1DM and HLA was first reported in 1973, following observation of increased frequency of HLA antigens in T1DM patients, compared with those in controls [19]. The HLA is located on chromosome 6p21 and encodes class I and class II genes. Class I genes, including HLA-A, HLA-B, and HLA-C are at the telomeric proximal end of the locus, whereas class II genes, including HLA-DP, HLA-DQ, and HLA-DR are more centromeric [20].
The HLA region accounts for approximately 50% heritability of T1DM. Specifically, HLA-DR and HLA-DQ are strongly associated with T1DM [21]. Haplotypes of class II molecules present antigens that can remarkably increase or decrease the binding ability of related auto-antigens, contributing to development of T1DM [22]. The highest risk haplotypes belong to HLA class II: DR4-DQA1*03:01-DQB1*03:02 (also referred to as DR4-DQ8) and HLA-DRB1*03:01-DQA1*05:01-DQB1*02:01 (also referred to as DR3-DQ2) (Table 1). Approximately 90% of patients with T1DM carry either DR4-DQ8 or DR3-DQ2, compared to 40% of the general population that do not have either of the haplotypes [23]. Approximately 30% of patients with T1DM have both of the haplotypes (DR3/4 heterozygotes), compared with 2% of the population that have both haplotypes but do not have T1DM. Siblings that share both HLA haplotypes (DR3/4-DQ8) with their diabetic probandsibling have higher risks for islet autoimmunity (85% by age 15), compared with siblings who do not (20% by age 15) [24]. Interaction between these 2 haplotypes, which gives rise to the DR4-DQ8/DR3-DQ2 genotype, confers the highest risk for T1DM, with an average odds ratio of 16 [25]. The DR3, DRB4, and DRB5 alleles are also associated with a risk of T1DM [26]. In contrast, some haplotypes have protective effects. For example, DQB1*06:02-DRB1*15:01-DQA1*01:02 (also referred to as DR2) is detected in approximately 20% of the population but in only 1% of T1DM individuals (odds ratio, 0.03) [27].
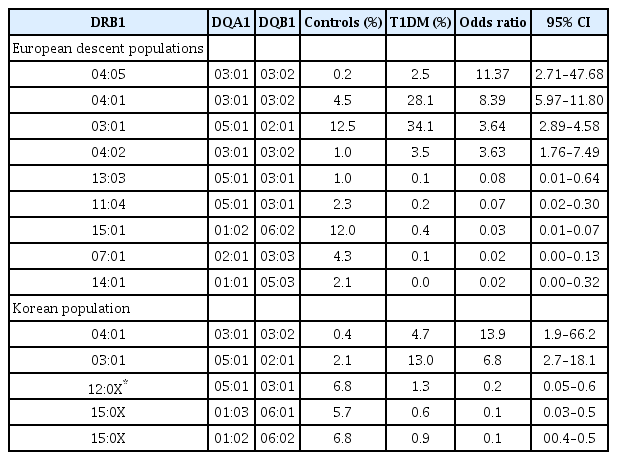
HLA class II DR-DQ genotypes and type 1 diabetes mellitus (T1DM) susceptibility in Caucasians and Koreans
HLA class I (HLA-A, HLA-B, and HLA-C) genes are associated with T1DM pathogenesis due to their interaction with HLA class II genes, although the risk for T1DM in patients with HLA class I haplotypes is relatively low compared to those with HLA-DR and -DQ [28]. Specifically, HLA-B*57:01 was identified as a protective variant against T1DM (odds ratio, 0.19), while HLA-B*39:06 increased its risk (odds ratio, 10.31) [29]. Some studies have shown that HLA-DPB1 can also influence T1DM susceptibility via modulating HLA class II-mediated risk, independent of HLA-DRB1 and -DQB1 [30]. Additionally, the HLA class III locus contains genes that are involved in immunological responses, which include the MHC class I chain-related gene A (MICA), tumor necrosis factor-alpha, and complement protein genes. The MICA gene polymorphism contributes to the risk of T1DM development and is associated with the age of disease onset [31].
Non-HLA genes associated with T1DM
Candidate gene approaches and GWAS studies in international and national groups have made significant progress in discovery of genes or loci associated with the risk of T1DM [13]. Several studies have reported that more than 50 additional loci are associated with T1DM [32-34]. The insulin gene (INS), on chromosome 11p15.5, is strongly associated with T1DM. There is a variable number of tandem repeats (VNTR) within the 5'-untranslated regions of the coding sequence of INS known as INS-VNTR. Several studies have reported that INS-VNTR may play a significant role in regulating insulin expression [35]. The number of repeats within sequenced alleles ranges from 26 to >200, with three classes of alleles identified based on overall size: class I alleles (26 to 63 repeats), class II alleles (64 to 143 repeats), and class III alleles (140 to 210 repeats) [36]. Class I VNTR alleles are associated with a predisposition to T1DM (odds ratio >2), while class III VNTR alleles are usually considered as protective against T1DM [37].
Polymorphism (rs2476601) of protein tyrosine phosphatase non-receptor type 22 (PTPN22) is also associated with T1DM [38]. The PTPN22 gene on chromosome 1p13 encodes lymphoid specific phosphatase which acts as a suppressor of T-cell activation. PTPN22 is crucial for maintaining balance between host defense and self-tolerance [39]. Bottini et al. [40] reported that the heterozygous variant (rs2476601) of PTPN22 was present in 30.6% of T1DM patients; however, it was only present in 21.3% of normal controls, thus increasing the risk of T1DM by approximately 2-fold. This polymorphism contributes to susceptibility to T1DM by increasing negative regulation of T-cell activation.
The cytotoxic T-lymphocyte antigen (CTLA-4) gene on chromosome 2q33 is another T1DM susceptibility gene and it encodes a T-cell specific transmembrane co-receptor. CTLA-4 binds to ligand B7 and acts as a negative regulator of cytotoxic T cells by down-regulating interleukin-2 receptor expression [41,42]. In a meta-analysis study, polymorphisms, CT60A/G (rs3087243) and +49A/G (rs231775), correlated with a greater risk of T1DM (odds ratio, 1.31 and 1.47, respectively) [43]. Although the exact mechanism by which the CTLA-4 gene affects autoimmune diseases is unclear, it may possibly be related to alteration of post-transcriptional regulation [44].
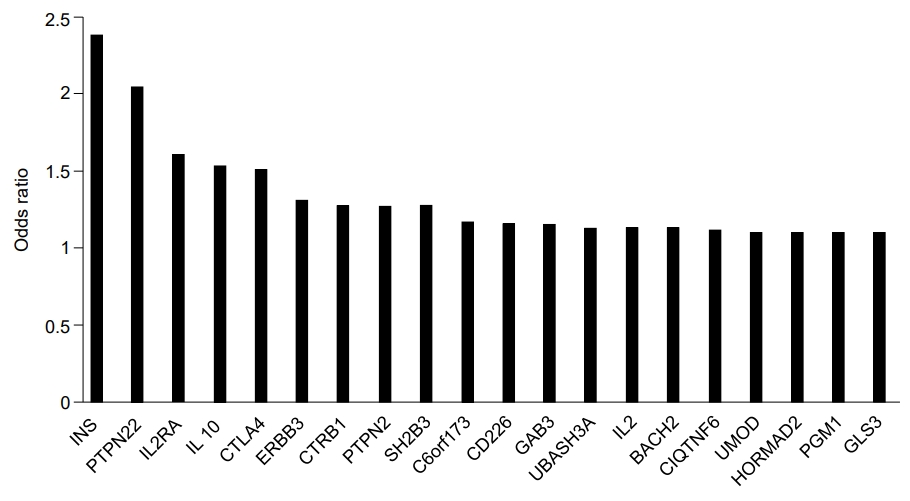
Other genes that reportedly influence the risk of T1DM include interleukin-2 receptor subunit alpha (IL2RA), interleukin genes (mainly IL-4 and IL-13), protein tyrosine phosphatase, non-receptor type 2 (PTPN2), interferon-induced helicase (IFIH1), basic leucine zipper transcription factor 2 (BACH2), Gli-similar 3 protein (GLIS3), and ubiquitin-associated and SH3 domain-containing protein A (UBASH3A) (Fig. 1) [32]. IL2RA variants affect the sensitivity of IL-2, an essential component for T-regulatory cell function, thus increasing the risk of T1DM [45]. IL-4 and IL-13 function to modulate autoimmunity. Furthermore, the IL-4/IL-13 pathway contributes to maintenance of peripheral tolerance and restrains development of T1DM [46]. PTPN2 is highly expressed in immune-related cells and its expression is modified in CD4+ T cells and more than 10 genetic regions in PTPN2 that demonstrate susceptibility to T1DM have been identified [47]. Reports have indicated that IFIH1, a cytoplasmic sensor of viral double-stranded RNA (dsRNA), mediates induction of the interferon response to viral RNA [48]. BACH2 has been observed as a modulator of innate immunity, antiviral responses, and activation of apoptosis in pancreatic beta-cells [49]. Additionally, GLIS3 is related to insulin expression and secretion of betacell and INS expression [50]. UBASH3A is reported to inhibit the NF-kB signaling pathway, resulting in reduced IL-2 gene expression [51].
Prediction of islet autoimmunity and T1DM
The association between genetics and T1DM is increasingly being recognized and elucidated. Many T1DM-associated variants associated with relatively high odds ratios have been discovered since the advent of GWAS [52]. Early identification of a high risk for T1DM in individuals based on genetics will allow intervention before islet autoimmunity is triggered. Genetic risk scores (GRS) or polygenic risk scores are widely used for predicting incidence and progression of complex genetic diseases. GRS may be particularly useful in addressing the current challenge of translating vast knowledge pertaining to T1DM genetics into clinically usable information for personalization of risks, diagnosis, and therapeutic options [53].
In a DAISY study, researchers followed high risk siblings, that were offspring of T1DM patients, and newborns from the population with high risk HLA genes [54]. They found that PTPN22, UBASH3A, and HLA-DR/DQ genotypes were associated with islet autoimmunity and T1DM risk. Forty-five percent of the study population with a combination of UBASH3A AA (rs11203203) and HLA-DR, as well as all subjects with PTPN22 TT (rs2476601), developed T1DM by age 15, compared to 3% of children with all other genotypes [55]. In the TrialNet Pathway to Prevention study, 1,244 subjects that were relatives of diabetic individuals or had at least one positive autoantibody were genotyped. The results indicated that a higher GRS, which can be determined using HLA single nucleotide polymorphisms (SNPs) and 30 non-HLA SNPs, significantly increased the rate of T1DM progression, after adjusting for confounding factors [9]. Winkler et al. [56] developed a weighted genetic score with a set of 40 SNPs that have been reported to be associated with T1DM in first-degree relatives using multivariate logistic regression in the BABYDIAB study. The weighted risk model, developed with selected SNPs, significantly improved T1DM prediction. The GRS for T1DM is also used to distinguish between T1DM and T2D or monogenic diabetes in youth (MODY). In the WTCCC study (n=3,887), GRS determined by using 30 SNPs, including HLA and non-HLA loci, discriminated between T1DM and MODY or monogenic neonatal diabetes [57].
Genetic susceptibility to T1DM in Koreans
Studies on the genetics of T1DM in non-Caucasian populations are relatively rare, especially in Korea, and possible differences in genetic variants of T1DM in East Asians and Caucasians have been recognized [58]. Jung et al. [59] analyzed HLA and non-HLA T1DM associated variants in 50 Korean T1DM patients. They found that frequencies of HLA-DRB1*04, DRB1*09, and DQB1*04 in T1DM patients increased significantly compared to those in healthy controls. In comparison, HLA-DRB1*14, DRB1*15, DQB1*05, and DQB1*06 showed a protective effect. In another study, the effects of DRB1-DQB1 haplotypes on T1DM susceptibility were similar in Koreans and Caucasians [60]. The distribution of the polymorphism +49A/G (rs231775) in the CTLA-4 gene in Korean T1DM patients and controls were not different [61]. Park et al. [62] analyzed polymorphisms in MICA exon 5 in 119 Korean T1DM patients and found that a MICA microsatellite allele that consists of 4 repetitions (A4) was positively associated with T1DM, while 6 repetitions of GCT/AGC (A6) demonstrated a protective effect. Chung et al. [63] analyzed the INS-VNTR in 352 Korean patients with T1DM. A class I genotype was found in 97.4% of patients and the frequency was higher than in the Caucasian population (nearly 80%) [64].
Conclusion
T1DM is characterized by autoimmune destruction of pancreatic islet cells. For decades, many studies have indicated that genetics play a crucial role in the etiology and pathogenesis of T1DM. GWAS and genome-wide linkage analysis have recently made significant contributions to current knowledge on the impact of genetics on T1DM development and progression. In this review, the HLA gene complex and non-HLA genes, such as INS, PTPN22, and CTLA-4 were associated with the development and progression of TID. However, at present, integrating and applying the wealth of knowledge related to T1DM genetics to diagnosis and prevention is challenging. An accurate T1DM predictive model based on genetic studies would allow intervention in at-risk individuals well before islet autoimmunity is triggered. Additionally, considering the influence of ethnic differences relative to incidence of T1DM, future studies are needed to identify T1DM-associated variants specific to the Korean population.
Notes
Conflicts of interest
No potential conflict of interest relevant to this article was reported.