 |
 |
- Search
| Ann Pediatr Endocrinol Metab > Volume 21(4); 2016 > Article |
|
Abstract
Nuclear receptors are in general ligand-dependent transcription factors that control a variety of mammalian physiologies including development, differentiation, proliferation, and homeostasis. Recent studies have found that two nutrient-sensing nuclear receptors, peroxisome proliferator-activated receptor ╬▒ and farnesoid x receptor, responding to fasting or feeding state, respectively are able to regulate autophagy, an evolutionarily conserved catabolic process involved in lysosomal degradation. In this review, we discuss the role of these nutrient-sensing nuclear receptors in an aspect of transcriptional regulation of autophagy, and how these nuclear receptor-driven transcriptional programs integrate lipophagy, a lipid autophagy with fatty acid oxidation to coordinate hepatic lipid metabolism in the fasted state of the liver.
The incidence of nonalcoholic fatty liver disease (NAFLD) and nonalcoholic steatohepatitis (NASH) is a global pandemic particularly in developed countries probably due to western diets and sedentary lifestyle1). NAFLD and NASH also occur in children whose prevalence of prediabetes and type 2 diabetes is significantly increased2).
For the past 30 years, the diverse members of nuclear receptors (NRs) have a great impact on metabolic diseases3,4,5). Of particular, activation of nutrient-sensing NRs with their synthetic ligands has beneficial effects on the improvement of these pathogeneses6,7,8). However, a detailed understanding of molecular mechanisms by which controlling NRs treat metabolic disorders in liver and other tissues is not fully established. Here, we discuss a novel function of nutrient-sensing NRs, which exert the transcriptional control of autophagy, a self-eating process conserved in all eukaryotic cells. We first discuss a basic concept of NRs followed by the metabolic roles of 2 nutrient-sensing NRs peroxisome proliferator-activated receptor ╬▒ (PPAR╬▒ or NR1C1) and farnesoid x receptor (FXR or NR1H4). We then shift our focus to autophagy with an emphasis of macroautophagy. We finally discuss how nutrient-sensing NRs regulate macroautophagy, highlighting an integration of lipophagy, a lipid autophagy with fatty acid oxidation (FAO) to satisfy energy demands during starvation.
As ligand-activated transcription factors (TFs), NRs serve as an interface between cellular or whole body environmental changes and our genome, providing an important link between transcriptional regulation and physiology. In particular, NRs play important functions in metazoan intercellular signaling because they converge diverse intra- and extracellular signals for initiating transcriptional programs for their relevant physiologies. Because of these unique roles in mammalian physiology, the activities of NRs are typically affected by a variety of environmental factors that function as ligands themselves or modulate ligand production5,9,10). Although there are a few exceptions, NRs are usually composed of several domains: an N-terminal ligand-independent activation function 1 motif, a DNA-binding domain (DBD) with 2 zinc finger motifs, a flexible hinge domain, and a C-terminal ligand-binding domain (LBD) consisting of 12 ╬▒-helixes. Among them, the helix 12 corresponds to ligand-dependent activation function 2 motif, which undergoes a conformational change upon an agonist binding to the ligand-binding pocket of NR. This provides a docking site for coactivators, which lead to the recruitment of more transcriptional machinery proteins to initiate the transcription of a given NR target gene5). The human genome encodes 48 members of NRs that include classical endocrine receptors for steroid hormones, thyroid hormone, and fat soluble vitamins and their derivatives, adopted orphan receptors for fatty acids (FAs), phospholipids, cholesterol metabolites, and bile acids (BAs), and orphan receptors whose ligands have not been identified yet or may not exist at all5).
The comprehensive and regulatory functions of NRs on the target genes have led to intense research on these TFs, contributing to enhancing our understanding of complex molecular mechanisms of transcriptional biology. Therefore, NRs have a tremendous impact on almost every mammalian physiology at all levels of cells, tissues, organs and an entire organism. They are also tightly associated with many human diseases including cancer, cardiovascular diseases, innate and adaptive immune disorders, neurological diseases, and metabolic diseases10,11). Moreover, NRs are often considered as valuable therapeutic targets to effectively treat human disease, which provides opportunities for developing better synthetic ligands with fewer side effects.
Among 48 members of NRs in human genome, PPAR╬▒ and FXR are the most prominent NRs whose activities are regulated by the availability of nutrients or fasting-feeding cycle, which plays a critical role in maintaining energy homeostasis in metabolic tissues including the liver.
The finding that a synthetic chemical Wy-14,643 treatment to rodents dramatically increases the numbers of peroxisomes in the liver leads to the discovery of PPAR╬▒, a master TF for lipid metabolism under conditions of energy deprivation or fasting11,12). Physiologic or pharmacologic activation of PPAR╬▒ by fasting or synthetic ligands markedly promotes uptake, utilization, and breakdown of FAs via upregulation of genes involved in FA transport, FA binding, peroxisomal and mitochondrial ╬▓-oxidation, and ketogenesis9,10,11). Moreover, PPAR╬▒ also induces the expression of fibroblast growth factor 21 (Fgf21) gene, an endocrine and/or autocrine hormone with pleiotropic effects in many metabolic tissues, which show antiobesity and antidiabetic effects13,14,15,16,17,18). As a ligand-activated NR, FAs such as arachidonic acid and polyunsaturated FAs, and C16:0-18:1 phosphatidylcholine have been reported to be endogenous agonist ligands for PPAR╬▒, which is also markedly activated by synthetic fibrate derivatives, a potent hypolipidemic drugs for patients with dyslipidemia6,19,20). Fenofibrate, one of fibrate derivatives, effectively lowers serum triglycerides, which are applied to the treatment of coronary heart disease and NAFLD4,6). PPAR╬▒ is highly expressed in tissues requiring effective FAO such as liver, brown adipose tissue, heart, skeletal muscle, kidney, intestine, and adrenal gland. Knockout studies of Ppar╬▒ gene in mice also demonstrate that PPAR╬▒ is absolutely required for proper lipid catabolism in the fasted state of the liver21,22). PPAR╬▒ forms heterodimeric complex with RXR, which typically binds to a direct repeat 1 response element (DR1 RE, 5' AGGTCA N AGGTCA 3', N; any single nucleotide) in the regulatory region of target genes10,11).
FXR is a BA NR that controls BA homeostasis and its associated lipid and glucose metabolism, and that is required for normal liver regeneration and may mediate the beneficial effect of vertical sleeve gastrectomy in mice23,24,25,26). Chenodeoxycholic acid (CDCA), one of BA species has been found to be an endogenous agonist ligand for FXR in major metabolic tissues such as the liver and intestine27). It is generally believed that in these tissues FXR is activated by CDCA derived from an enterohepatic BA circulation process, suggesting that FXR potently responds to a fed state of the liver28). Therefore, FXR plays an essential role in BA homeostasis by remarkably suppressing hepatic expression of Cyp7a1 gene encoding cholesterol 7 ╬▒-hydroxylase, a rate-limiting enzyme of BA synthesis. FXR is also pharmacologically regulated by several synthetic or natural ligands such as obeticholic acid (OCA, also known as 6╬▒-ethyl-CDCA or INT-747), GW4064, fexaramine, guggulsterone, and cafestol29,30,31,32,33). Moreover, BA- or pharmacologic FXR activation in the intestine is sufficient for inducing the expression of fibroblast growth factor 15 (Fgf15) gene, an endocrine hormone participating in a part of the negative feedback regulations of BA synthesis, and in subsequent elevations of proteins and glycogen synthesis in the liver34,35). As did PPAR╬▒, FXR also forms a heterodimeric receptor complex with RXR, which predominantly binds to an inverted repeat 1 response element (IR1 RE, 5' AGGTCA N TGACCT, N; any single nucleotide) in the regulatory region of many target genes. These transcriptional regulations of FXR target genes have a critical impact on the physiology of the fed state. Finally, US federal drug administration has recently approved the treatment of OCA, a semisynthetic CDCA analogue to patients with primary biliary cirrhosis, which might pave the way for diverse applications to other metabolic and/or inflammatory diseases36,37).
Autophagy, a self-eating process throughout all eukaryotic cells is a fundamentally conserved degradation mechanism involving lysosomal delivery of cargo molecules such as various soluble materials, membrane-enclosed organelles, and even invasive parasites38,39). Although there is a basal autophagic activity in most cells and/or tissues, autophagy is highly inducible by numerous stimuli including physiologic stresses (e.g., nutrient deprivation, hypoxia, high temperature, high density condition, exercise, etc.), hormones (e.g., glucagon, etc.), pharmacologic reagents (e.g., rapamycin, Torin 1, etc.), or many disease conditions (e.g., cancer, myopathy, etc.)40). Although there are several types of autophagy depending on classifying criteria, macroautophagy is a major type of autophagy and have been most intensively studied so far compared to other types of autophagy. When the macroautophagy (herein referred to as autophagy) occurs, an isolated membrane called a phagophore sequesters cytoplasmic components to form double-membrane vesicle called an autophagosome (AG). AG then fuses with the lysosome to become an autolysosome (AL) where the autophagic cargos are degraded by acidic lysosomal hydrolases. It is also necessary to note that AL can be generated by a fusion of lysosome with amphisome, a vesicle created from a fusion between AG and late endosome (also called multivesicular body)41). Subsequently, biological building blocks (e.g., amino acids, FAs, nucleotides, and glucose) degraded from macromolecules or organelles within ALs are released into cytoplasm, which is recycled to synthesize new macromolecules or used for energy supplementations. In this view, autophagy itself acting as a cellular degradation process substantially links catabolism into anabolism39,42). The catabolic function of autophagy is also associated with the quality control of many intracellular components by eliminating misfolded or unfolded proteins, or worn-out organelles. Therefore, cellular functions of autophagy are very diverse, ranging from eliminating superfluous organelles to providing amino acids and ATPs for energy supplementation and new protein synthesis to removing aggregate prone proteins for a quality control mechanism. Autophagy also plays an essential role in destroying invasive pathogens and subsequently presenting pathogen-derived antigens on the plasma membrane38). Finally too much autophagy seems to trigger certain types of cell death including apoptosis43). Beyond its cellular functions, autophagy has a broad impact on mammalian physiology and pathology including embryonic development, innate and adaptive immunity, neurodegenerative disease, cancer, heart disease and skeletal pathogenesis, ageing, metabolic disease, and so on44). On the contrary of a previous concept, suggesting that autophagy be a nonselective catabolic process, autophagy can degrade very selective cargo molecules and organelles. The latter includes proteins (aggrephagy), glycogen (glycophagy), lipid droplets (lipophagy), iron-bearing ferritins (ferritinophagy), the ribosome (ribophagy), the peroxisome (pexophagy), the endoplasmic reticulum (reticulophagy), the mitochondria (mitophagy), virus and bacteria (xenophagy), and so forth45,46).
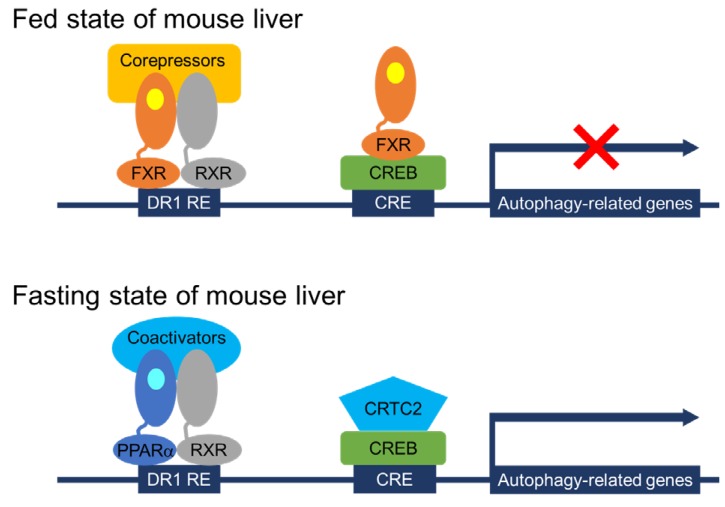
It has been relatively well known that acute regulation of autophagy by nutrient-sensing pathways is largely dependent on upstream signaling molecules including the mechanistic target of rapamycin complex 1 (mTORC1) and other kinases47). However, a longer-term regulation of autophagy has not been fully established yet. Particularly, transcriptional and epigenetic regulation of autophagy has been recently appreciated48,49). Lee et al.50) and Seok et al.51) have shown that hepatic autophagy is controlled by several TFs including two nutrient-sensing NR PPAR╬▒ and FXR, and cAMP responsive element binding protein (CREB), a well-known fasting activated TF for hepatic gluconeogenesis (Fig. 1). Both PPAR╬▒ and CREB activated by fasting status induce hepatic autophagy via a direct upregulation of core autophagy-related genes, which also lead to lipophagy, one of selective autophagies, resulting in the release of free FAs from lipid droplets52,53,54). It is of interest to note that PPAR╬▒ is not only important for inducing FAO and ketogenesis to provide ATPs and alternative fuels for the brain, but also for increasing lipophagy to supply free FAs as substrates for the FAO during fasted state of the mammalian liver11,19). It would be very intriguing to identify novel PPAR╬▒ target genes encoding lipophagy adaptor proteins.
In contrast, the BA receptor FXR represses hepatic autophagy by at least 2 mechanisms (Fig. 1). One mechanism proposed by Lee et al.50) is that FXR directly suppresses the expression of autophagy-related genes via a genomic competition with PPAR╬▒ binding to DR1 RE that is often found in the regulatory regions of many autophagy-related genes. Consistent with this finding, previous studies have already demonstrated that FXR activation is able to repress ApoCIII and ApoA expression by binding to DR1 RE in the promoters of both genes where FXR may act as ligand-dependent transrepressor rather than transactivator55,56). This mechanism is further supported by a comprehensive analysis of hepatic PPAR╬▒ and FXR cistromes, showing their remarkable overlapping peaks containing DR1 RE throughout the whole mouse genome, which strongly suggests a possibility that both NRs compete for binding to shared sites in the regulatory regions of core autophagy-related genes, with opposite transcriptional outputs. On the other hand, the other mechanism proposed by Seok et al.51) is similar but distinct from that of Lee et al.50). They have shown that the piggyback binding of FXR to CREB without DNA binding in the promoter regions of autophagy-related genes is able to dissociate and subsequently expel CRTC2, a well-known coactivator from the nucleus. This leads to turning off the expression of many autophagy-related genes including TFEB, a master TF for the lysosomal biogenesis57). Previous studies by the Ballabio laboratory have shown that TFEB acts as a transcriptional activator for lysosomal and autophagy-related genes, which lead to the induction of autophagy. It is of interest to note that the catabolic effects of TFEB also depend on the induction of PGC1╬▒ and the presence of PPAR╬▒, suggesting a complementary function of TFEB-PGC1╬▒-PPAR╬▒ axis during fasting58). These results uncover complex but complementary genomic circuits in which transcriptional programs controlled by PPAR╬▒ and CREB-CRTC2 induce hepatic autophagy, which is then markedly suppressed by FXR activation. Transcriptional circuits of autophagy-related genes governed by several TFs further integrate autophagy with long-term physiologic nutrient responses48,49,59). These studies also suggest that beneficial effects of fibrate derivatives or OCA on patients with hyperlipidemia or primary biliary cirrhosis, respectively could be attributed to coordinating hepatic autophagy in addition to their known biochemical pathways, and that controlling autophagy by targeting PPAR╬▒ and FXR might provide a novel therapeutic strategy for the pathogenesis of a wide range of human diseases.
It has been shown that PPAR╬▒ and FXR, responding to fasting or feeding state, respectively are fundamental nutrient-sensing NRs, which orchestrate proper programs of transcription involved in FAO, ketogenesis, or BA homeostasis. The findings of Lee et al.50) and Seok et al.51) have extended the roles of these NRs to autophagy regulation. Although the details of underlying mechanisms by which PPAR╬▒ and FXR counteract each other for autophagy regulation still need to be further elucidated, transcriptional regulation of autophagy by both NRs seems to be to some extent mTORC1-independent. It would also be of interest to identify direct PPAR╬▒ or FXR target genes encoding key enzymes, which can modulate posttranslational modifications (PTMs) of core autophagy machinery proteins. Intriguingly, both mechanisms proposed by Lee et al.50) and Seok et al.51) are somewhat similar but still quite distinct from each other, suggesting that there should be complex transcriptional circuits governing mammalian autophagy, and that the expression of autophagy-related genes should be complementarily coordinated by several TFs including nutrient-sensing NRs, CREB, and TFEB. Taken together, nutrient deprivation allows mammalian cells to perform elaborate signaling pathways to rapidly trigger autophagy initiation by changing PTMs of autophagy machineries and to provide autophagy machinery proteins themselves by inducing the expression of autophagy-related genes. The latter may lead to the preparation of much longer-term of nutrient deprivation in the liver. Finally, investigations need to be performed to determine if the models proposed by Lee et al.50) and Seok et al.51) also control autophagy in other metabolic tissues, and if these mechanisms account for therapeutic effects of targeting PPAR╬▒ or FXR on metabolic liver diseases.
Acknowledgments
We apologize for contributors whose work was not cited due to limited space. This research was supported by Kyungpook National University Research Fund, 2015.
References
1. Sayiner M, Koenig A, Henry L, Younossi ZM. Epidemiology of nonalcoholic fatty liver disease and nonalcoholic steatohepatitis in the united states and the rest of the world. Clin Liver Dis 2016;20:205ŌĆō214. PMID: 27063264.


2. Newton KP, Hou J, Crimmins NA, Lavine JE, Barlow SE, Xanthakos SA, et al. Prevalence of prediabetes and type 2 diabetes in children with nonalcoholic fatty liver disease. JAMA Pediatr 2016;170:e161971. PMID: 27478956.



3. Wagner M, Zollner G, Trauner M. Nuclear receptors in liver disease. Hepatology 2011;53:1023ŌĆō1034. PMID: 21319202.


4. Shulman AI, Mangelsdorf DJ. Retinoid x receptor heterodimers in the metabolic syndrome. N Engl J Med 2005;353:604ŌĆō615. PMID: 16093469.


5. Sonoda J, Pei L, Evans RM. Nuclear receptors: decoding metabolic disease. FEBS Lett 2008;582:2ŌĆō9. PMID: 18023286.



6. Shipman KE, Strange RC, Ramachandran S. Use of fibrates in the metabolic syndrome: A review. World J Diabetes 2016;7:74ŌĆō88. PMID: 26981181.



7. Trivedi PJ, Hirschfield GM, Gershwin ME. Obeticholic acid for the treatment of primary biliary cirrhosis. Expert Rev Clin Pharmacol 2016;9:13ŌĆō26. PMID: 26549695.


8. Ding L, Pang S, Sun Y, Tian Y, Yu L, Dang N. Coordinated actions of FXR and LXR in metabolism: from pathogenesis to pharmacological targets for type 2 diabetes. Int J Endocrinol 2014;2014:751859. PMID: 24872814.




9. Chawla A, Repa JJ, Evans RM, Mangelsdorf DJ. Nuclear receptors and lipid physiology: opening the X-files. Science 2001;294:1866ŌĆō1870. PMID: 11729302.


10. Evans RM, Mangelsdorf DJ. Nuclear Receptors, RXR, and the Big Bang. Cell 2014;157:255ŌĆō266. PMID: 24679540.



11. Evans RM, Barish GD, Wang YX. PPARs and the complex journey to obesity. Nat Med 2004;10:355ŌĆō361. PMID: 15057233.


12. Issemann I, Green S. Activation of a member of the steroid hormone receptor superfamily by peroxisome proliferators. Nature 1990;347:645ŌĆō650. PMID: 2129546.



13. Inagaki T, Dutchak P, Zhao G, Ding X, Gautron L, Parameswara V, et al. Endocrine regulation of the fasting response by PPARalpha-mediated induction of fibroblast growth factor 21. Cell Metab 2007;5:415ŌĆō425. PMID: 17550777.


14. Owen BM, Bookout AL, Ding X, Lin VY, Atkin SD, Gautron L, et al. FGF21 contributes to neuroendocrine control of female reproduction. Nat Med 2013;19:1153ŌĆō1156. PMID: 23933983.



15. Zhang Y, Xie Y, Berglund ED, Coate KC, He TT, Katafuchi T, et al. The starvation hormone, fibroblast growth factor-21, extends lifespan in mice. Elife 2012;1:e00065. PMID: 23066506.



16. Owen BM, Ding X, Morgan DA, Coate KC, Bookout AL, Rahmouni K, et al. FGF21 acts centrally to induce sympathetic nerve activity, energy expenditure, and weight loss. Cell Metab 2014;20:670ŌĆō677. PMID: 25130400.



17. Talukdar S, Owen BM, Song P, Hernandez G, Zhang Y, Zhou Y, et al. FGF21 regulates sweet and alcohol preference. Cell Metab 2016;23:344ŌĆō349. PMID: 26724861.



18. Kliewer SA, Mangelsdorf DJ. Fibroblast growth factor 21: from pharmacology to physiology. Am J Clin Nutr 2010;91:254SŌĆō257S. PMID: 19906798.



19. Bocos C, G├Čttlicher M, Gearing K, Banner C, Enmark E, Teboul M, et al. Fatty acid activation of peroxisome proliferator-activated receptor (PPAR). J Steroid Biochem Mol Biol 1995;53:467ŌĆō473. PMID: 7626496.


20. Chakravarthy MV, Lodhi IJ, Yin L, Malapaka RR, Xu HE, Turk J, et al. Identification of a physiologically relevant endogenous ligand for PPARalpha in liver. Cell 2009;138:476ŌĆō488. PMID: 19646743.



21. Lee SS, Pineau T, Drago J, Lee EJ, Owens JW, Kroetz DL, et al. Targeted disruption of the alpha isoform of the peroxisome proliferator-activated receptor gene in mice results in abolishment of the pleiotropic effects of peroxisome proliferators. Mol Cell Biol 1995;15:3012ŌĆō3022. PMID: 7539101.



22. Kersten S, Seydoux J, Peters JM, Gonzalez FJ, Desvergne B, Wahli W. Peroxisome proliferator-activated receptor alpha mediates the adaptive response to fasting. J Clin Invest 1999;103:1489ŌĆō1498. PMID: 10359558.



23. Kalaany NY, Mangelsdorf DJ. LXRS and FXR: the yin and yang of cholesterol and fat metabolism. Annu Rev Physiol 2006;68:159ŌĆō191. PMID: 16460270.


25. Huang W, Ma K, Zhang J, Qatanani M, Cuvillier J, Liu J, et al. Nuclear receptor-dependent bile acid signaling is required for normal liver regeneration. Science 2006;312:233ŌĆō236. PMID: 16614213.


26. Ryan KK, Tremaroli V, Clemmensen C, Kovatcheva-Datchary P, Myronovych A, Karns R, et al. FXR is a molecular target for the effects of vertical sleeve gastrectomy. Nature 2014;509:183ŌĆō188. PMID: 24670636.



27. Tu H, Okamoto AY, Shan B. FXR, a bile acid receptor and biological sensor. Trends Cardiovasc Med 2000;10:30ŌĆō35. PMID: 11150726.


28. Zhou H, Hylemon PB. Bile acids are nutrient signaling hormones. Steroids 2014;86:62ŌĆō68. PMID: 24819989.



29. Pellicciari R, Fiorucci S, Camaioni E, Clerici C, Costantino G, Maloney PR, et al. 6alpha-ethyl-chenodeoxycholic acid (6-ECDCA), a potent and selective FXR agonist endowed with anticholestatic activity. J Med Chem 2002;45:3569ŌĆō3572. PMID: 12166927.


30. Maloney PR, Parks DJ, Haffner CD, Fivush AM, Chandra G, Plunket KD, et al. Identification of a chemical tool for the orphan nuclear receptor FXR. J Med Chem 2000;43:2971ŌĆō2974. PMID: 10956205.


31. Downes M, Verdecia MA, Roecker AJ, Hughes R, Hogenesch JB, Kast-Woelbern HR, et al. A chemical, genetic, and structural analysis of the nuclear bile acid receptor FXR. Mol Cell 2003;11:1079ŌĆō1092. PMID: 12718892.



32. Urizar NL, Liverman AB, Dodds DT, Silva FV, Ordentlich P, Yan Y, et al. A natural product that lowers cholesterol as an antagonist ligand for FXR. Science 2002;296:1703ŌĆō1706. PMID: 11988537.


33. Ricketts ML, Boekschoten MV, Kreeft AJ, Hooiveld GJ, Moen CJ, M├╝ller M, et al. The cholesterol-raising factor from coffee beans, cafestol, as an agonist ligand for the farnesoid and pregnane X receptors. Mol Endocrinol 2007;21:1603ŌĆō1616. PMID: 17456796.



34. Inagaki T, Choi M, Moschetta A, Peng L, Cummins CL, McDonald JG, et al. Fibroblast growth factor 15 functions as an enterohepatic signal to regulate bile acid homeostasis. Cell Metab 2005;2:217ŌĆō225. PMID: 16213224.


35. Kir S, Beddow SA, Samuel VT, Miller P, Previs SF, Suino-Powell K, et al. FGF19 as a postprandial, insulin-independent activator of hepatic protein and glycogen synthesis. Science 2011;331:1621ŌĆō1624. PMID: 21436455.




36. Rinella ME. Nonalcoholic fatty liver disease: a systematic review. JAMA 2015;313:2263ŌĆō2273. PMID: 26057287.


37. Bowlus CL. Obeticholic acid for the treatment of primary biliary cholangitis in adult patients: clinical utility and patient selection. Hepat Med 2016;8:89ŌĆō95. PMID: 27621676.



38. Mizushima N, Komatsu M. Autophagy: renovation of cells and tissues. Cell 2011;147:728ŌĆō741. PMID: 22078875.


40. Mizushima N, Yoshimori T, Levine B. Methods in mammalian autophagy research. Cell 2010;140:313ŌĆō326. PMID: 20144757.



41. Gordon PB, Seglen PO. Prelysosomal convergence of autophagic and endocytic pathways. Biochem Biophys Res Commun 1988;151:40ŌĆō47. PMID: 3126737.


42. Galluzzi L, Pietrocola F, Levine B, Kroemer G. Metabolic control of autophagy. Cell 2014;159:1263ŌĆō1276. PMID: 25480292.



43. Mari├▒o G, Niso-Santano M, Baehrecke EH, Kroemer G. Self-consumption: the interplay of autophagy and apoptosis. Nat Rev Mol Cell Biol 2014;15:81ŌĆō94. PMID: 24401948.



44. Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature 2008;451:1069ŌĆō1075. PMID: 18305538.



45. Jin M, Liu X, Klionsky DJ. SnapShot: selective autophagy. Cell 2013;152:368ŌĆō368.e2. PMID: 23332767.



46. Stolz A, Ernst A, Dikic I. Cargo recognition and trafficking in selective autophagy. Nat Cell Biol 2014;16:495ŌĆō501. PMID: 24875736.


47. Inoki K, Kim J, Guan KL. AMPK and mTOR in cellular energy homeostasis and drug targets. Annu Rev Pharmacol Toxicol 2012;52:381ŌĆō400. PMID: 22017684.


48. F├╝llgrabe J, Ghislat G, Cho DH, Rubinsztein DC. Transcriptional regulation of mammalian autophagy at a glance. J Cell Sci 2016;129:3059ŌĆō3066. PMID: 27528206.


49. F├╝llgrabe J, Klionsky DJ, Joseph B. The return of the nucleus: transcriptional and epigenetic control of autophagy. Nat Rev Mol Cell Biol 2014;15:65ŌĆō74. PMID: 24326622.


50. Lee JM, Wagner M, Xiao R, Kim KH, Feng D, Lazar MA, et al. Nutrient-sensing nuclear receptors coordinate autophagy. Nature 2014;516:112ŌĆō115. PMID: 25383539.



51. Seok S, Fu T, Choi SE, Li Y, Zhu R, Kumar S, et al. Transcriptional regulation of autophagy by an FXR-CREB axis. Nature 2014;516:108ŌĆō111. PMID: 25383523.



52. Singh R, Kaushik S, Wang Y, Xiang Y, Novak I, Komatsu M, et al. Autophagy regulates lipid metabolism. Nature 2009;458:1131ŌĆō1135. PMID: 19339967.



53. Singh R, Cuervo AM. Lipophagy: connecting autophagy and lipid metabolism. Int J Cell Biol 2012;2012:282041. PMID: 22536247.




54. Liu K, Czaja MJ. Regulation of lipid stores and metabolism by lipophagy. Cell Death Differ 2013;20:3ŌĆō11. PMID: 22595754.



55. Chennamsetty I, Claudel T, Kostner KM, Baghdasaryan A, Kratky D, Levak-Frank S, et al. Farnesoid X receptor represses hepatic human APOA gene expression. J Clin Invest 2011;121:3724ŌĆō3734. PMID: 21804189.



56. Claudel T, Inoue Y, Barbier O, Duran-Sandoval D, Kosykh V, Fruchart J, et al. Farnesoid X receptor agonists suppress hepatic apolipoprotein CIII expression. Gastroenterology 2003;125:544ŌĆō555. PMID: 12891557.


57. Sardiello M, Palmieri M, di Ronza A, Medina DL, Valenza M, Gennarino VA, et al. A gene network regulating lysosomal biogenesis and function. Science 2009;325:473ŌĆō477. PMID: 19556463.


58. Settembre C, De Cegli R, Mansueto G, Saha PK, Vetrini F, Visvikis O, et al. TFEB controls cellular lipid metabolism through a starvation-induced autoregulatory loop. Nat Cell Biol 2013;15:647ŌĆō658. PMID: 23604321.



59. Settembre C, Di Malta C, Polito VA, Garcia Arencibia M, Vetrini F, Erdin S, et al. TFEB links autophagy to lysosomal biogenesis. Science 2011;332:1429ŌĆō1433. PMID: 21617040.



Fig.┬Ā1
Working mechanisms of transcriptional regulation of autophagy by nutrient-sensing NRs and CREB. In the fed state of wild-type mice, FXR activated by its endogenous agonist CDCA (shown in yellow colored circle) binds to DR1 RE as a heterodimeric complex with RXR in the distal regulatory regions of autophagy-related genes. Moreover, activated FXR can also form a piggyback interaction with a CRE-bound CREB in the proximal regulator regions of autophagy-related genes via the dissociation of a coactivator CRTC2. These 2 different mechanisms may account for the transcriptional repression of autophagy-related genes by FXR activation in the fed state of mouse liver. In contrast, fasting-activated PPAR╬▒ by its endogenous ligands, FFAs or PC (16:0-18:1)(shown in light blue colored circle), binds to DR1 RE via the formation of a heterodimeric complex with RXR in the distal regions of autophagy-related genes. CREB also recruits its coactivator CRTC2 in the proximal regions of autophagy-related genes. These complimentary transcription complexes ensure that fasting leads to turning on the expression of many core autophagy-related genes in the fasted state of mouse livers. Overall, activated PPAR╬▒ or FXR competes with each other for binding to shared DNA sequences in the distal regions of autophagy-related genes. Additionally, FXR competes with CRTC2 for binding to CREB in the proximal regions of autophagy-related genes. NRs, nuclear receptors; CDCA, chenodeoxycholic acid; FFAs, free fatty acids; PC, phosphatidylcholine; FXR, farnesoid x receptor; RXR, retinoid x receptor; DR1 RE, direct repeat 1 response element; CRE, cAMP response element; CREB, cAMP response element binding protein; PPAR╬▒, peroxisome proliferator-activated receptor ╬▒; CRTC2, CREB regulated transcription coactivator 2.
- TOOLS
- Related articles in APEM







